NIGMS is committed to supporting a wide-ranging portfolio of biomedically relevant fundamental research. As we discussed in a previous Feedback Loop post, we see this approach as the best way to increase our understanding of life. For many years, one important dimension of diversity in our scientific portfolio—the organisms scientists use to conduct their research—was limited by technical considerations. However, recent advances such as the decreasing cost of genome sequencing and the development of the CRISPR system for genetic modification now make it possible to use an expanded range of research organisms.
Applying these new technologies to the broader universe of Earth’s species, some of which have been the subject of research for many years and some of which have only recently attracted academic attention, presents an opportunity for a fresh perspective on the nature and behavior of living systems. In recognition of this opportunity, and as an extension of the recent portfolio analysis of NIH support for a subset of traditional model organisms presented by NIH’s Office of Extramural Research (OER), we decided to explore NIGMS’ support for investigator-initiated research using a subset of organisms for which historical application numbers are low. The 17 research organisms listed in Table 1 below were suggested for analysis by NIGMS program staff, who encountered them as the subject of one or more applications to NIGMS since 2008. On average, the number of applications per organism was never greater than three per year. Although this is not meant to be a comprehensive list of the rare research organisms supported by NIGMS, it spans the phylogenetic tree and is thus representative.
keywords
Myxococcus xanthus
“Rhodospirillum centenum” OR “Rhodospirillum rubrum”
“Methanococcus” OR “maripaludis”
“Zoothamnium” OR “giant ciliate”
“Echinoidea” OR “Strongylocentrotus purpuratus” OR “sea urchin”
“Cnidaria” OR “Hydra”
“Euprymna scolopes” OR “bobtail squid”
Nasonia
“Tribolium castaneum” OR “red flour beetle”
“Ciona intestinalis” OR “Ciano savignyi” OR “sea squirt”
“Ginglymostoma cirratum” OR “nurse shark”
“Myxini” OR “Hagfish”
Lamprey
“Takifugu rubripes” OR “Japanese puffer” OR “Tiger puffer” OR “pufferfish” OR “puffer fish”
“Ambystoma mexicanum” OR “axolotl”
Anolis carolinensis OR “Anolis carolinesis” OR “Carolina anole”
“Taeniopygia guttata” OR “zebra finch” OR “zebrafinch”
Applications were first identified by searching the text of the entire NIGMS portfolio of Research Project Grant (RPG—defined here as R01, R37, R35, DP2, R15, R24, R21, and R00 mechanisms) applications from Fiscal Years (FY) 2008 through 2016 for keywords associated with each of the 17 species (Table 1). Each application identified in this manner was read by at least two curators to confirm the use of the research organism in question. If the initial coding did not produce a consensus, the discrepancy was resolved by further discussion. This manual curation approach is consistent with the methodology employed in the above-mentioned analysis of traditional research organisms presented by OER, so the two datasets could be compared.
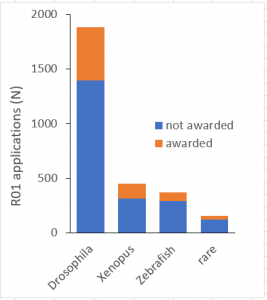
Figure 1 shows the number of R01 applications received and awards made by NIGMS to study the widely used research organisms Drosophila melanogaster, Xenopus laevis/tropicalis, and Danio rerio (Zebrafish) from FY 2008 to 2015; in comparison, the use of our selected 17 research organisms in R01 applications over the same time period has been relatively rare (n = 152). Consistent with existing data on trans-NIH award rates, NIGMS R01 applications that propose to study the widely used models enjoy a higher combined award rate than the overall average (Table 2; Fisher’s exact p-value < 0.0001).
Although the sample size is small, R01 applications proposing to study rare research organisms have an award rate similar to applications on Drosophila, Xenopus, and Zebrafish or the overall NIGMS R01 pool (Table 2; Fisher’s exact p-value = 0.13 and 0.50, respectively). This conclusion, which may seem contrary to expectations, is unchanged if data on applications through the end of FY 2016 and/or data on other RPG mechanisms are included in the calculation of award rates.
| applications | awards | award rate | |
| all NIGMS | 28429 | 6513 | 22.9% |
| Drosophila, Xenopus, Zebrafish | 2701 | 704 | 26.1% |
| rare research organisms | 152 | 31 | 20.4% |
While NIGMS continues to support the use of traditional research organisms, we also welcome applications using new and unusual research organisms that propose well-justified studies relevant to the Institute’s mission. As we go forward, we will continue to monitor the distribution of support for the various organisms studied by NIGMS grantees, including how those organisms may be linked to particular areas of study. As always, we are interested to hear your thoughts on this issue, and encourage PIs who are interested in applying to NIGMS to contact a program director who manages applications close to their area of research.
We are grateful to the NIH Office of Portfolio Analysis in the Division of Program Coordination, Planning, and Strategic Analysis for sharing their data on Drosophila, Xenopus, and Zebrafish awards. We would also like to thank our colleagues on the NIGMS Research Organisms Working Group as well as Michael Bender, Dylan Burgoon, and Donna Krasnewich for their help with this analysis.
Jake Basson, Office of Program Planning, Analysis and Evaluation
Emily Carlson, Office of Communications and Public Liaison*
Lisa Dunbar, Office of Scientific Review
Rafael Gorospe. Division of Research Capacity Building
Sailaja Koduri, Division of Training, Workforce Development, and Diversity
Paul Sammak, Division of Biophysics, Biomedical Technology, and Computational Biosciences
Kristine Willis, Division of Genetics and Molecular, Cellular, and Developmental Biology
Dorit Zuk, Division of Genetics and Molecular, Cellular, and Developmental Biology


Interesting analysis. I note that the lack of curated model organism databases for
these organisms may be an impediment to scientific progress, particularly using high throughput techniques.