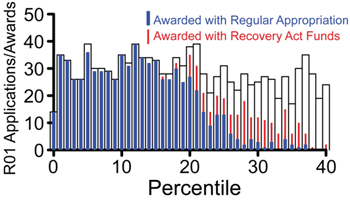
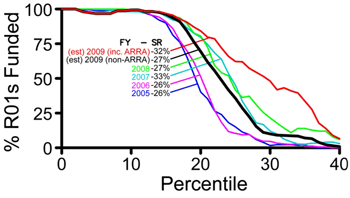
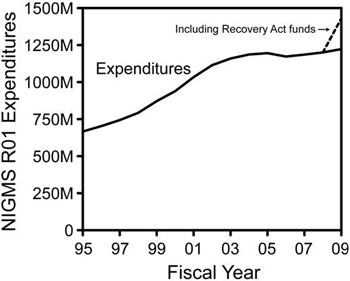
The Collaborative Research for Molecular and Genomic Studies of Basic Behavior in Animal Models program announcement (PA-07-096) will expire on January 8, 2010. We will be reissuing this program announcement in 2010, but not in time for you to submit an application for the February or March R01 deadlines.
The purpose of PA-07-096 and its successor is to facilitate collaborative research between investigators with expertise in animal behavior and those with expertise in molecular biology and/or genomics that addresses questions about the mechanisms of behavior in animal models. The long-term goal is to develop new or enhanced animal models for studying aspects of behavior relevant to the NIH mission. We encourage applications from multiple PIs.
The first deadlines for applications submitted in response to the reissued program announcement will be June 7, 2010 (for new applications) and July 6, 2010 (for resubmissions). Beginning in June and July 2010, we’ll start accepting applications for the standard R01 deadlines: June/July, October/November and February/March.
Regardless of when you apply or resubmit an application for this reissued program announcement, you’ll need to use the new R01 application forms and instructions with shorter page limits.
Curious about what the reissued announcement will look like? Ask me!