Greetings from Lindau, Germany, where I and my NIH colleagues, Irene Eckstrand and Katrin Eichelberg, are attending the 61st Lindau Nobel Laureate Meeting  . This year’s meeting, focused on physiology and medicine, has brought together 23 Nobel laureates and 566 outstanding pre- and postdoctoral students, including 80 from the United States, to share their passion for science and their commitment to finding solutions to the world’s biggest problems.
. This year’s meeting, focused on physiology and medicine, has brought together 23 Nobel laureates and 566 outstanding pre- and postdoctoral students, including 80 from the United States, to share their passion for science and their commitment to finding solutions to the world’s biggest problems.
This is the third year that NIGMS has participated in the Lindau program, and the first year in which participation was NIH-wide.
Each morning, six Nobel laureates have given short lectures about the history of their science, their successes and failures, and their visions of the future. Among the NIGMS-funded laureates giving these talks are Elizabeth Blackburn, Oliver Smithies, Thomas Steitz, Roger Tsien and Ada Yonath. The afternoons are reserved for free-ranging discussions among the laureates and young researchers. It has been a delight to watch the two groups get to know each other and discuss important scientific problems of global interest and importance.
On the opening day, Countess Bettina Bernadotte, whose family has sponsored the meeting since its beginning, welcomed the assembly. Her enthusiasm for science and her commitment to the meetings was apparent in her introduction of the Lindau mission – “Educate. Inspire. Connect” – and her advice that we “never cease to be curious.” Expanding on this, Annette Schavan, the German Federal Minister of Education and Research, stressed that connections among the generations, such as those made at this meeting, are crucial to scientific progress.
The American delegation, which was sponsored by NIH, the Department of Energy, Oak Ridge Associated Universities and Mars Incorporated, organized a U.S.-themed International Day on Monday, June 27. This series of events truly jumpstarted the week.
As part of International Day, NIH-supported Nobel laureate Peter Agre explained his view of the scientist-citizen who uses his or her knowledge and talents to make the world a better place. The discoverer of aquaporins, Agre talked about applying his research to fighting malaria in Zambia. NIGMS’ Irene Eckstrand then spoke about using the power of computing to inform policies for the control and eradication of infectious diseases. An energetic discussion ensued between the highly engaged students and speakers, focusing on efforts to understand the epidemiology of malaria, approaches to reducing the number of deaths from the disease and the importance of collaborative research between developed and developing nations.
The International Day evening program included a very well-received video address from NIH Director Francis Collins on the strength of American medical science. His message about NIH’s commitment to global health aligned perfectly with the focus of earlier talks, including a presentation by Bill Gates, who was inducted into the Honorary Senate of the Lindau Foundation.
Irene, Katrin and I are all quite proud to have been a part of this effort and look forward to sharing more about our experiences here.
Post written by Donna Krasnewich, Irene Eckstrand and Katrin Eichelberg (NIAID)



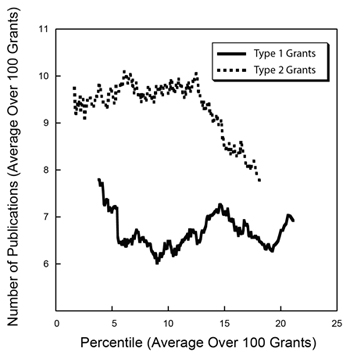
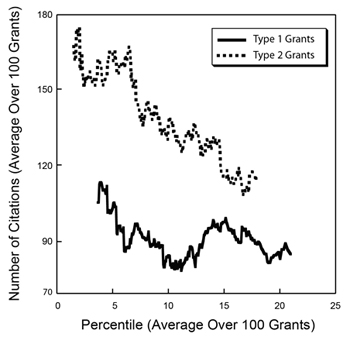
 As we anticipated last year, the
As we anticipated last year, the  As I enter my final few weeks at NIGMS, I’m engaged in a lot of transition planning. One major aspect is the designation of an acting director, and I’m happy to tell you that Judith Greenberg has agreed to serve in this capacity after my departure early next month. She was acting director in 2002 and 2003, after Marvin Cassman left and before I arrived, and I know that she will once again do a fantastic job.
As I enter my final few weeks at NIGMS, I’m engaged in a lot of transition planning. One major aspect is the designation of an acting director, and I’m happy to tell you that Judith Greenberg has agreed to serve in this capacity after my departure early next month. She was acting director in 2002 and 2003, after Marvin Cassman left and before I arrived, and I know that she will once again do a fantastic job.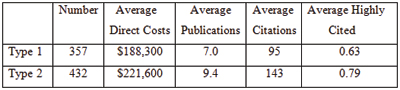

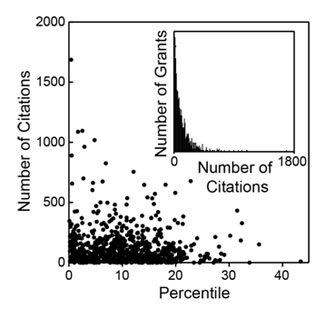
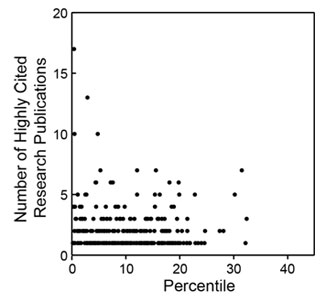
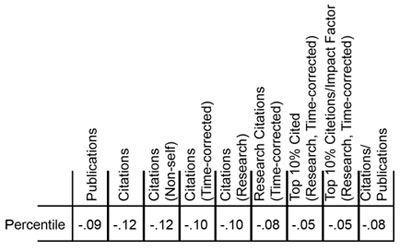
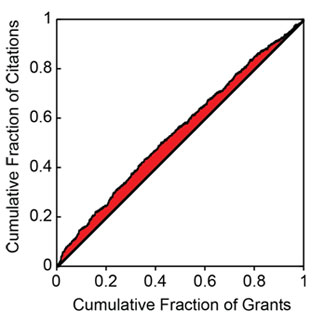
 Last week, NIGMS hosted the Frontiers in Cell Migration and Mechanotransduction meeting. It brought together an impressive group of scientists working at many levels, from molecules to cells, tissues and organs.
Last week, NIGMS hosted the Frontiers in Cell Migration and Mechanotransduction meeting. It brought together an impressive group of scientists working at many levels, from molecules to cells, tissues and organs.