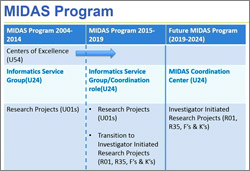
UPDATE: We thank the community for its initial feedback as we continue to develop plans for this program, which will support research within the NIGMS mission (including a limited number of clinical areas). The program will offer comparable levels of support as the program project (P01) mechanism, but its structure will be quite different. In addition to the capacity building and AIDS-Related Structural Biology program centers, we will continue to support the Biomedical Technology Research Resource centers (P41), Mature Synchrotron Resources (P30), and select coordinating or resource centers in areas of high strategic need.
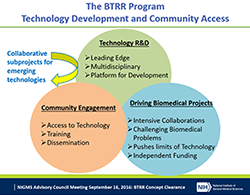
At its January 2017 meeting, our Advisory Council endorsed a concept for a new program to support collaborative, team-based science. This initiative is the result of evaluations of our previous programs, recent research on the science of team science  , and community input.
, and community input.
Many research questions in biomedical science can be pursued by single investigators and their close collaborators through single- or multi-principal investigator R01 grants. However, complex research questions may require the coordinated efforts of several research laboratories and closer collaborations among researchers with diverse areas of expertise. NIGMS recognizes the importance and benefits of supporting collaborative research teams when these are necessary to achieve important scientific breakthroughs or new understanding of phenomena.
NIGMS’ new Collaborative Program Grant is designed to support highly integrated, multidisciplinary research teams of three to six investigators who will address complex research questions, train and mentor new scientists, and impact scientific problems that would benefit from coordinated research support. The key application requirements are a single, integrated research program without subprojects and a multiple-principal investigator management plan. We expect to issue a funding opportunity announcement by the summer and, beginning in 2018, we will make four to six awards per year with annual direct costs ranging from $500,000 to $1.5 million. We plan to phase out our use of the P01 and most of our other center mechanisms. We will continue to support capacity building centers, such as those of the Institutional Development Award (IDeA) program, and to support the AIDS-Related Structural Biology program centers.
We encourage the community to watch the presentation at our council meeting, and we welcome your input and feedback on these plans. You can email your comments or post them here.